Monthly summary of sampled birds
The website will be updated regularly as more data from the project becomes available. Please check back for the latest information
May 2025 - Download here
April 2025 - Download here
March 2025 - Download here
February 2025 - Download here
January 2025 - Download here
December 2024 - Download here
November 2024 - Download here
The Nextstrain builds are available on the SENTINEL Wild Birds Group - https://nextstrain.org/groups/SentinelWildBirds
June 2025
1. OVERVIEW
SENTINEL Wild Birds aims to enhance the understanding of highly pathogenic avian influenza (HPAI) virus dynamics in wild bird populations by conducting active surveillance at key locations in and near Europe. These locations are divided into the following surveillance nodes: Node 1 Gulf of Finland (Finland, Estonia), Node 2 Southern Baltic Sea (Sweden, Latvia, Lithuania, Poland), Node 4 Eastern Black Sea (Georgia), Node 6 Lake Constance (Germany, Austria, Switzerland), Node 7 Veneto Region (Italy), Node 8 Camargue (France), and Node 9 Gulf of Cadiz (Spain). This monthly summary provides an update on sampled wild birds as part of an early warning system to support wildlife management and disease prevention efforts. The data in this report are based on previously unpublished samples collected from May to June 2025.
2. RESULT
2.1 DATA COLLECTION
Since the last monthly report (https://doi.org/10.5281/zenodo.15622783), and as of 15th June 2025, test results have been submitted for 44 samples taken from 26 individual wild birds representing 5 taxa across 2 nodes in Europe (Figure 1; Table 1 & 2). None of the samples were positive for avian influenza virus (Figure 1; Table 1 & 2). Samples from Node 7 consisted of cloacal, oropharyngeal and feather samples (all sample types from all birds), while samples from Node 2 consisted of combined oropharyngeal and cloacal samples.
TABLE 1 Total number of individuals sampled in the wild (including recaptures of some birds), as well as number of individuals tested positive for avian influenza virus in the respective country for the sampling period.
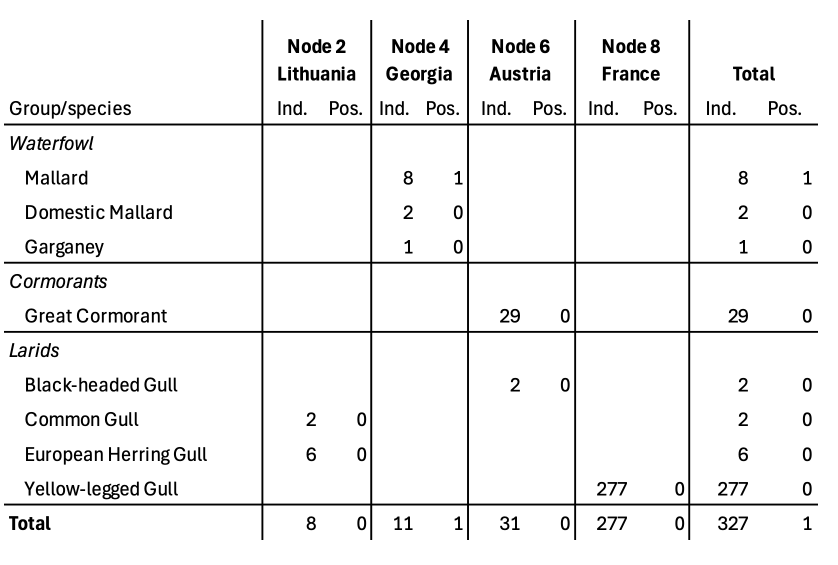
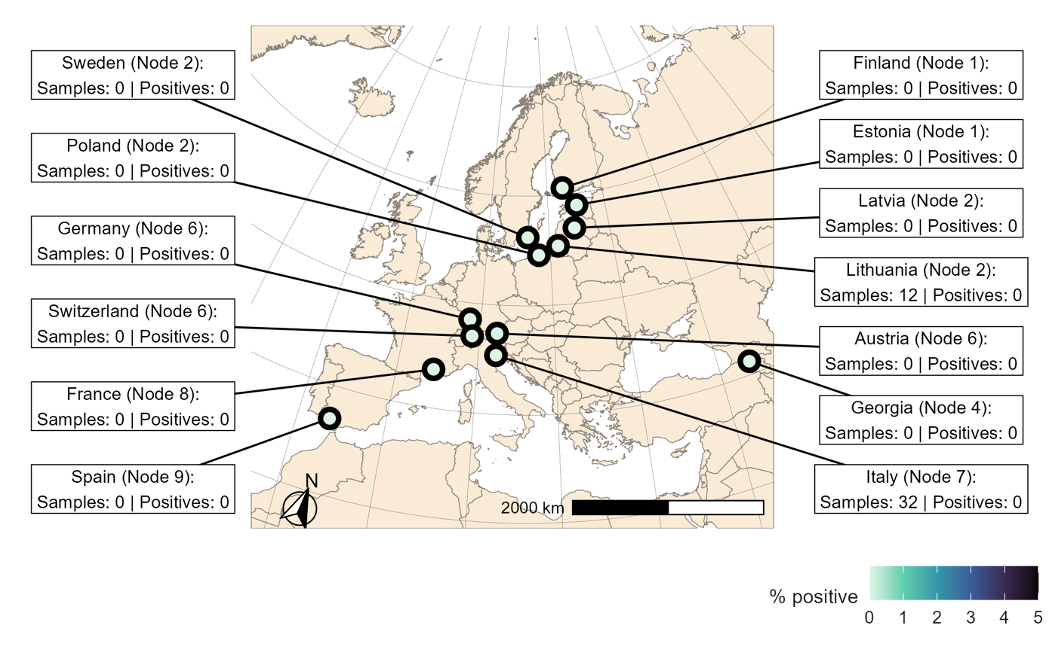
FIGURE 1 Sample sites for the birds sampled in during the reporting period.
TABLE 2 Total number of collected samples as well as number of samples positive for avian influenza virus, in the respective country during the sampling period.

2.2 GENOMICS SUMMARY
Since the last report was published (https://doi.org/10.5281/zenodo.15622783), and as of 15th June 2025, two new sequences have been generated from samples positive for H5 avian influenza virus. The samples were collected from a Eurasian Teal and a Mallard in France (Node 8, Camargue Region) and Spain (Node 9 Gulf of Cadiz), respectively. Both samples were H5N2, and genetic analysis of the haemagglutinin (HA) gene sequences found that both samples belonged to the Eurasian non-Goose Guangdong (EA-nonGsGd) low pathogenicity avian influenza (LPAI) clade.
The sequence from France (Node 8 Camargue Region) show similarity to other H5N2 and H5N3 EA-nonGsGd LPAI sequences from Sweden (Node 2 Southern Baltic Sea), as well as other sequences collected outside of the SENTINEL Wild Birds project from Europe and Asia. The sequence from Spain (Node 9 Gulf of Cadiz) also shows similarity to H5Nx EA-nonGsGd LPAI sequences from Europe, including two from Node 2 Southern Baltic Sea and Node 4 Eastern Black Sea, as well as other sequences collected outside of the project.
The Nextstrain builds are available on the SENTINEL Wild Birds Group (https://nextstrain.org/groups/SentinelWildBirds).
Apart from the H5 genome sequences, an additional seven non-H5 sequences were generated from samples collected in France (Node 8 Camargue Region; N=5) and Spain (Node 9 Gulf of Cadiz; N=2). The samples from France were of the following subtypes: 1xH2N3, 1xH6N9, 1xH7N6 and 2xH7N7, whilst the samples from Spain were of the H6N1 and H2N7 subtypes.
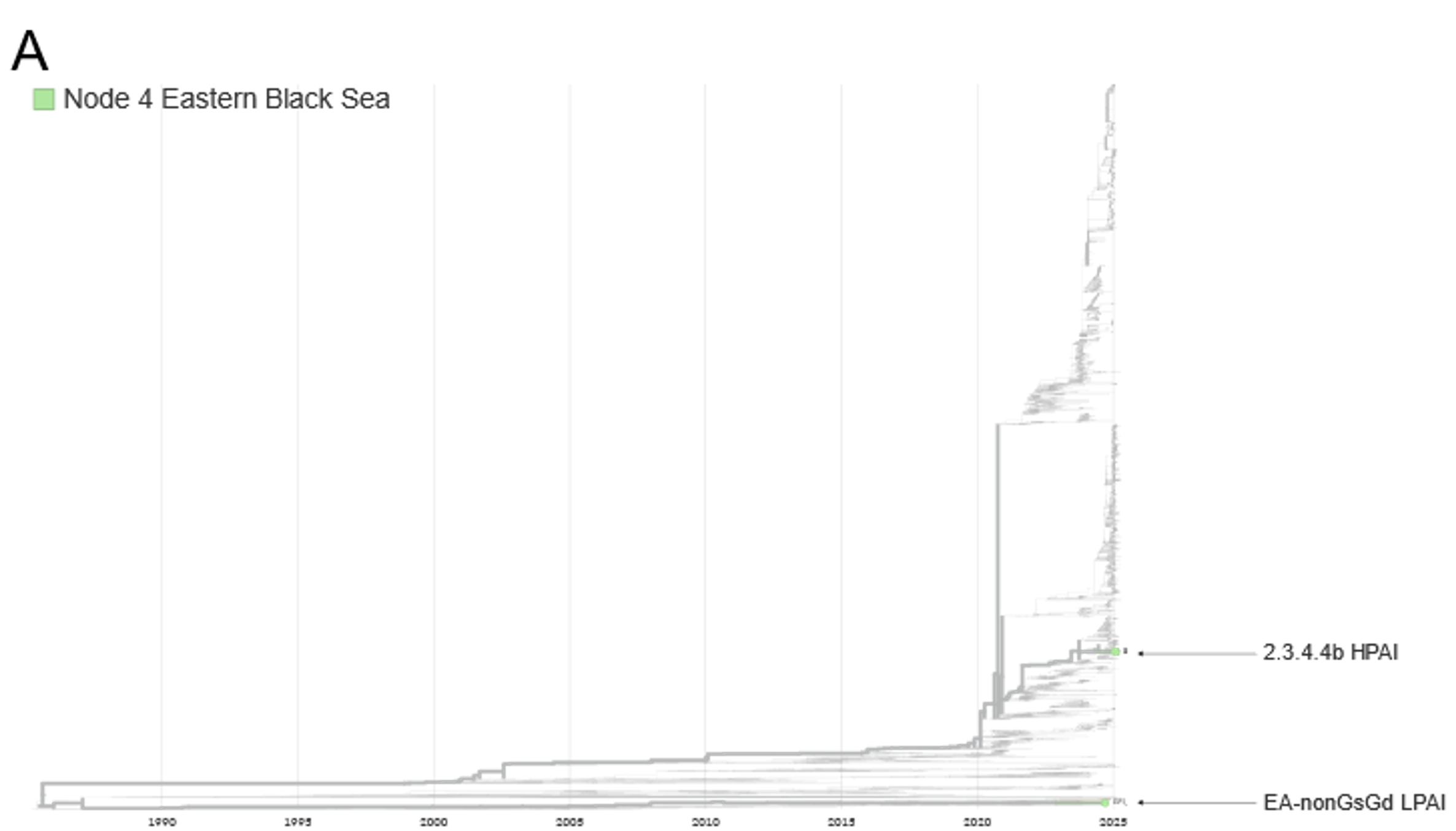
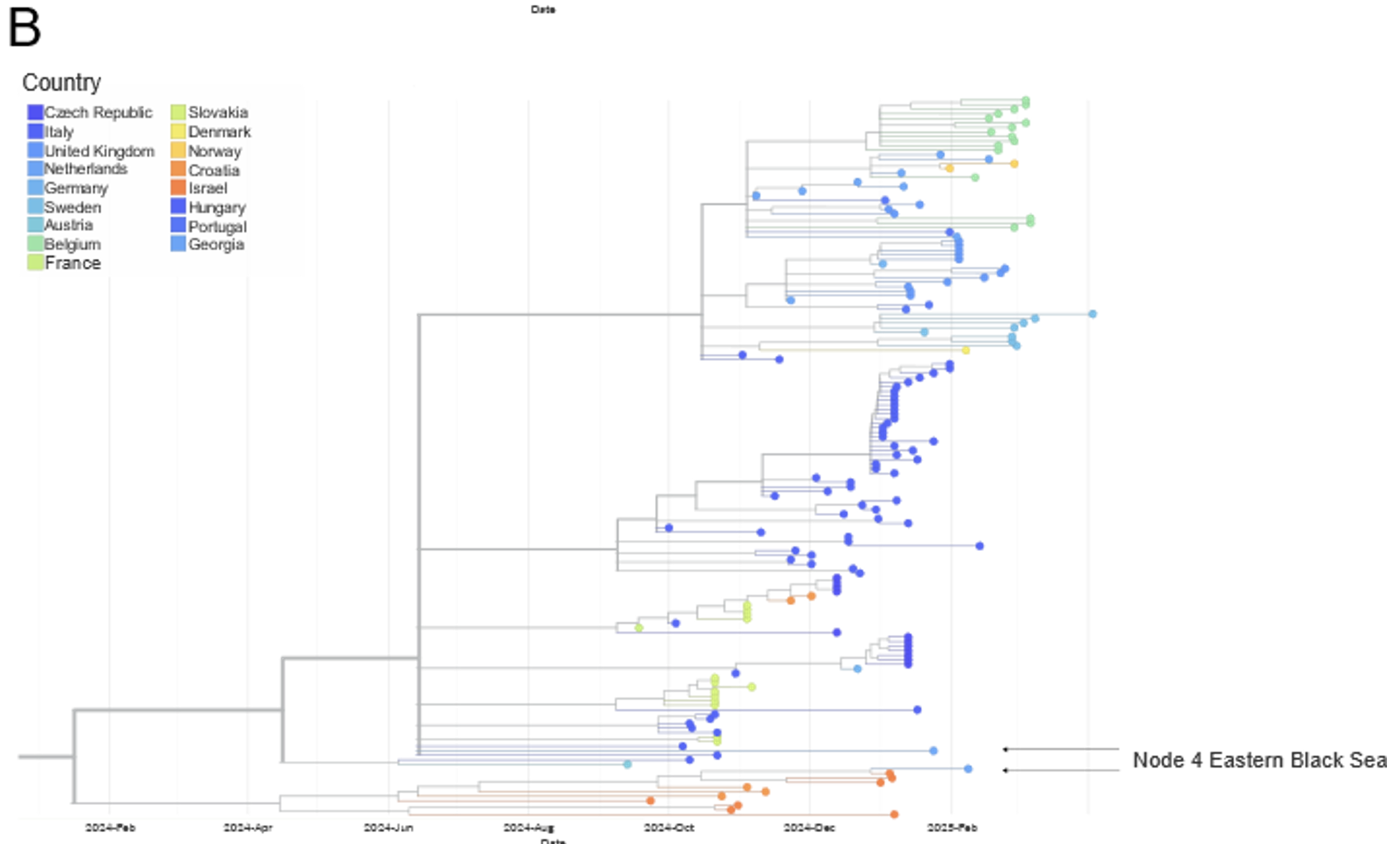
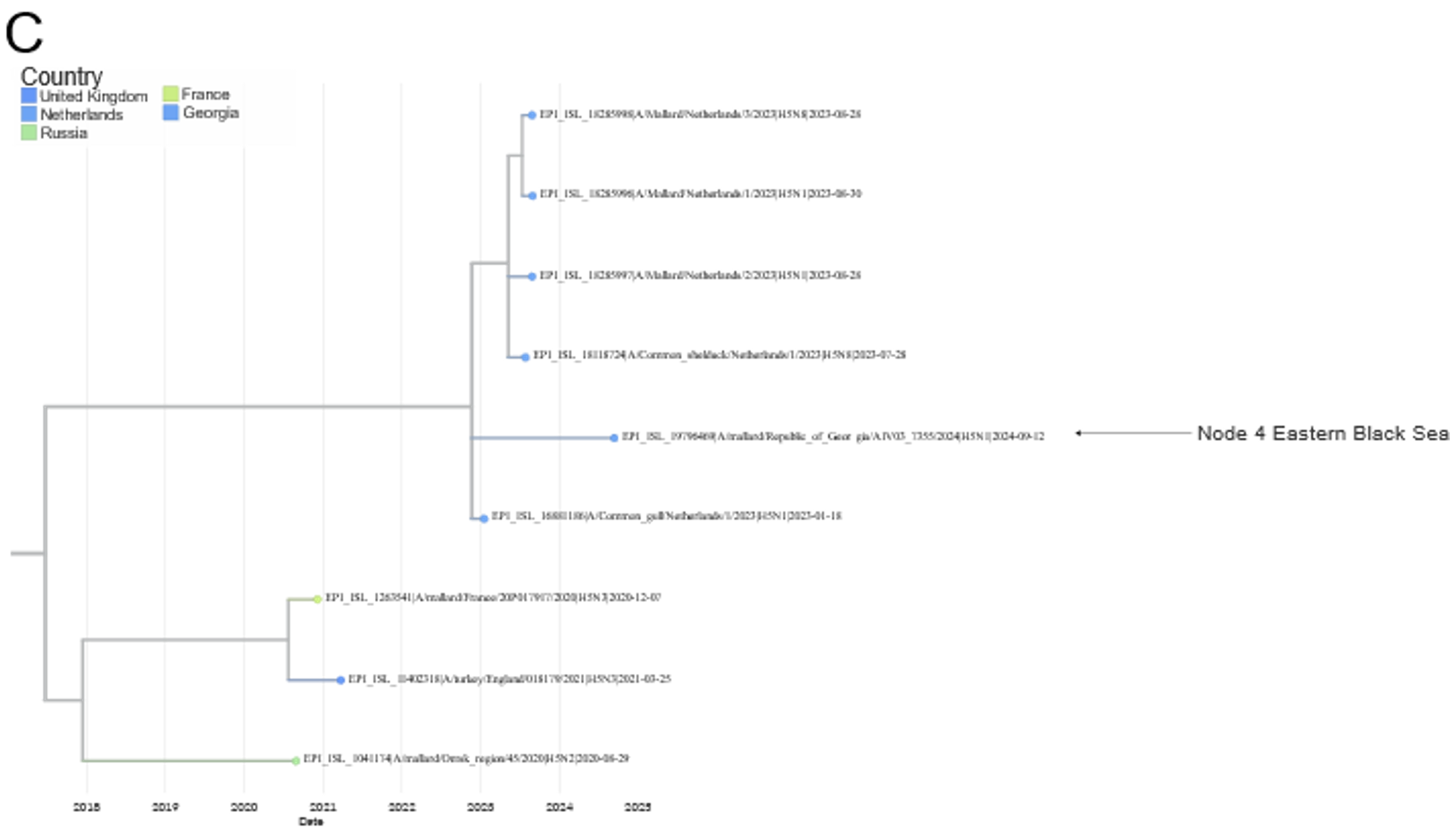
FIGURE 3 Phylogenetic analyses of the HA sequences generated by Node 8 Camargue Region and Node 9 Gulf of Cadiz. A. Image of the HA sequences from the EA-nonGsGd clade with the sequences from the SENTINEL Wild Bird nodes highlighted. B and C. Zoom images of the HA tree focusing on the sequences from Node 8 Camargue Region and Node 9 Gulf of Cadiz, respectively.
3. CONCLUSION
The SENTINEL Wild Birds project was designed to primarily detect viruses during migration and non-breeding periods of the year. This is reflected in this June report where a very limited number of samples were collected, and from only two nodes. All samples were negative. The sample size is too low to say anything conclusive about avian influenza circulation, but it fits general long-term datasets on low pathogenicity avian influenza virus in waterfowl that typically report low prevalence rates at this time of the year when birds breed and do not aggregate in large numbers. This is further evidenced by the generation of several low pathogenicity avian influenza genomes, from samples collected and reported in previous months. The H5 low pathogenicity avian influenza genomes in this report show similarity to those detailed in previous reports and continue to show connectivity between the sampling nodes of the SENTINEL Wild Birds project. In the last three years, Europe has experienced outbreaks of high pathogenicity influenza viruses in colonial breeding seabirds, such as gulls, terns, skuas and gannets, but this far in 2025 such outbreaks have been less frequent. In the coming months, we expect low numbers of sampled birds across most nodes, and also low numbers of samples positive for avian influenza.
ACKNOWLEDGMENT
This report is based on data collected and analysed by fieldworkers, laboratory personnel, and node coordinators from the following organizations and institutions:
Node 1: LABRIS, Riigi Laboriuuringute ja Riskihindamise Keskus (Estonia); Ruokavirasto, Finnish Food Authority (Finland); University of Turku (Finland)
Node 2: Swedish National Veterinary Institute (SVA) (Sweden); Linnaeus University (Sweden); Institute of Food Safety, Animal Health and Environment (BIOR) (Latvia); National Food and Veterinary Risk Assessment Institute (Lithuania); State Food and Veterinary Service (Lithuania); National Veterinary Research Institute (Poland)
Node 4: Centre of Wildlife Disease Ecology (CWDE), Ilia State University; State Laboratory of Agriculture of Georgia
Node 6: Austrian Agency for Health and Food Safety (AGES) (Austria); Verein für die Betreuung des Naturschutzgebietes Rheindelta (Naturschutzverein Rheindelta) (Austria); Friedrich-Loeffler-Institute (FLI) (Germany); Max-Planck-Institut für Verhaltensbiologie (MPI) (Germany); National Reference Centre for Poultry and Rabbit Diseases (NRGK) (Switzerland); Swiss Institute for Virology and Immunology (IVI) (Switzerland)
Node 7: Istituto Zooprofilattico Sperimentale delle Venezie; Istituto Superiore per la Protezione e la Ricerca Ambientale (ISPRA)
Node 8: École Nationale Vétérinaire de Toulouse (ENVT); INRAE (Institut National de Recherche pour l’Agriculture, l’Alimentation et l’Environnement); La Tour du Valat; Conservatoire d’Espaces Naturels d’Occitanie (CEN Occitanie); Laboratoire Départemental du Gard; Office Français de la Biodiversité (OFB); Ministère de l’Agriculture et de la Souveraineté Alimentaire; Muséum national d’Histoire naturelle (MNHN); Agence nationale de sécurité sanitaire de l’alimentation, de l’environnement et du travail (ANSES)
Node 9: Martina Ferraguti, Josué Martínez-de la Puente, and Jordi Figuerola at Estación Biológica de Doñana (EBD-CSIC); Ursula Höfle at Grupo de Sanidad y Biotecnología (SaBio), Instituto de Investigación en Recursos Cinegéticos (IREC-CSIC); Elisa Pérez-Ramírez and Jovita Fernández-Pinero at Centro de Investigación en Sanidad Animal (CISA-INIA-CSIC)




